Get up to date with the latest HitGen articles and join us in the events
While DNA-encoded library (DEL) selection has been applied to a variety of targets with multiple candidates progressed into different stages of drug development, the utilization of DEL in DNA- and RNA-binding proteins remains questionable. This is largely due to the fact that each DEL compound contains a long stretch of DNA barcode, which when used to screen against DNA- or RNA- binding proteins (transcription factors, DNA polymerases, helicases, DNases, RNases, chromatin modifying complexes, etc.) , would lead to significant false positives. To address this potential interference caused by DEL DNA tags, HitGen has made significant efforts in fine-tuning selection conditions as well as developing deconvolution algorithms. These practices, along with our trillion size DEL and 10+ year of DEL experience, have made it possible to conduct meaningful ligand discovery for DNA- or RNA-binding proteins.
DEL selection for transcription factors
Transcription factors (TFs) are involved in multiple biological processes and reflect a delicate system in gene expression regulation. TFs are generally considered as challenging targets because on one hand, most of them lack endogenous ligands (except for nuclear receptors); on the other hand, although most TFs bind to essential cofactors or DNA fragments, the interface of these interactions are generally large, flat and transient. In addition, the potential binding of DNA binding domains (DBD) to the DEL DNA tags further complicates DEL selection strategies. To alleviate these confounding factors, HitGen has employed an array of technologies such as using target proteins with different domains or mutations, counter targets, consensus DNA blocking in DEL selection (Figure 1).
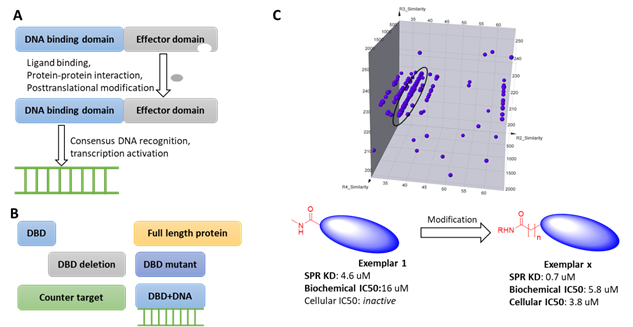
Figure 1, A) Schematic of transcription factor activation. B) Different strategies used in DEL selection. C) Representative signals and hit parameters.
DEL selection for DNA helicases
DNA helicases unwind DNA structures in an ATP-dependent manner and are essential for DNA replication and repair. Defects in DNA helicases can result in different genetic disorders in which genomic instability and predisposition to cancer are common features, warranting them as potential cancer treatment targets. However, the DNA binding nature of this family also presents potential hurdles of high background and false positives when employing DEL selection for their ligands. At HitGen, by implementing our customized selection strategies, we have successfully performed DEL selection for multiple helicase targets and identified different series of hits with good selectivity (Figure 2).
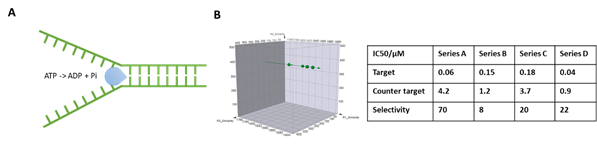
Figure 2, A) Schematic of DNA helicase function. B) Representative selection signals and hit parameters.
DEL selection for DNA polymerases
DNA polymerases create DNA molecules by assembling nucleotides in chain reactions. Due to their crucial functions in many DNA metabolic processes, multiple human diseases have been linked to mutations or defects of DNA polymerases, making them potential drug targets. In addition, inhibition on DNA polymerase also represents a good strategy on anti-microbial drug development. At HitGen, exquisitely designed screenings involving DNA substrates and different ligands have afforded multiple hits and yielded well-behaved drug candidates with good enzymatic inhibition activity and selectivity (Figure 3).
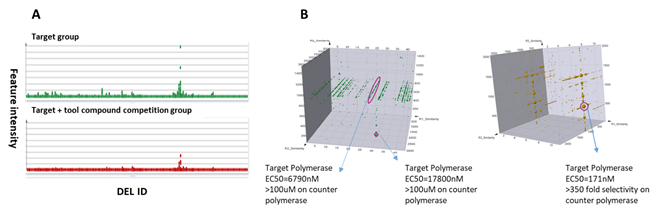
Figure 3, A) An example of screening output for a DNA polymerase target. B) Representative signals and hit parameters.
Computational approaches to find the needle in the haystack
In our pursuit of identifying small molecule ligands for DNA-binding proteins, it's crucial to discern the true positive signals from the noise introduced by the inherent affinity of these proteins for the DNA tags. At HitGen, we meticulously scrutinize the enrichment of DNA motifs from the Unique Molecule Identifier region (UMI region, which is a region with random bases) within the target samples, providing us with a comprehensive understanding of the overall DNA binding levels. Furthermore, we delve into the structure-signal relationship of the DEL molecules, enabling us to distinguish features that are enriched due to common DNA motifs, rather than pharmacophores on the DEL compounds. This rigorous process ensures us to minimize false positives and enhance the precision of our ligand discovery.
How about nucleases and RNA binding proteins?
Due to their DNA cleavage nature, DNA nucleases have been regarded non-compatible with DEL selection for a long time. With relentless efforts, HitGen is now able to conduct DEL selection for numerous nuclease targets by introducing proper mutations in those targets to abolish their DNA cleavage activities, and by depleting specific metal ions required for the reactions.
RNA helicases, RNases, RNA-dependent RNA polymerases, as well as other RNA processing/exporting proteins represent a large category of RNA-binding proteins. Although some RNA-binding proteins interact with DNA significantly, the binding affinity could be accurately assessed to evaluate the potential interference during DEL selection therefore presents less of a hurdle. At HitGen, we have successfully conducted multiple DEL screenings for various RNA-binding protein targets with validated hits identified.
DEL selection for DNA- and RNA-binding proteins at HitGen
HitGen is at the forefront of DEL design and screening, with numerous elegant strategies developed to tackle the most challenging targets. Contact us for DNA- and RNA-binding protein projects. Your chance of successful ligand development will be maximized with our demonstrated abilities:
●DNA binding compounds excluded from selection given the nature of DEL
●Comprehensive screening solutions to cover various target types
●Proprietary algorithms to double-check on DNA-binding false positives
●Proven track record of our screening experience and successful cases

HitGen Inc.
Building C2, NO.8, Huigu 1st East Road
Tianfu International Bio-Town,
Shuangliu District, Chengdu City,
Sichuan Province,P.R. China
Tel : +86-28-8519-7385
HitGen Pharmaceuticals Inc.(US Subsidiary)
Tel : +1-(508)-840-9646
Vernalis (R&D) Limited (UK Subsidiary)
Granta Park,Great Abington
Cambridge,CB21 6GB
United Kingdom
Tel : +44(0)1223-895555
 川公网安备 51012202000492号
川公网安备 51012202000492号
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information