Enabling Innovative Drug Discovery with Advanced Technologies
The human genome encodes more than 30,000 proteins taking roles in multiple biological functions yet the whole proteome landscape shows very limited space being represented as “druggable”. The discovery and exploration on the “undruggable” landscape has become an attractive pharmacological conundrum taking advantage of emerging techniques such as targeted protein degradation (TPD). TPD modulates protein target levels by hijacking the ubiquitin-proteasome system to induce degradation of the target, which, does not necessarily lead to inhibition of the target as the majority of drugs do. To facilitate the discovery of targeted protein degrader, HitGen has developed its DNA encoded library (DEL) technology to identify promising binders with known linkage point towards degradation targets.
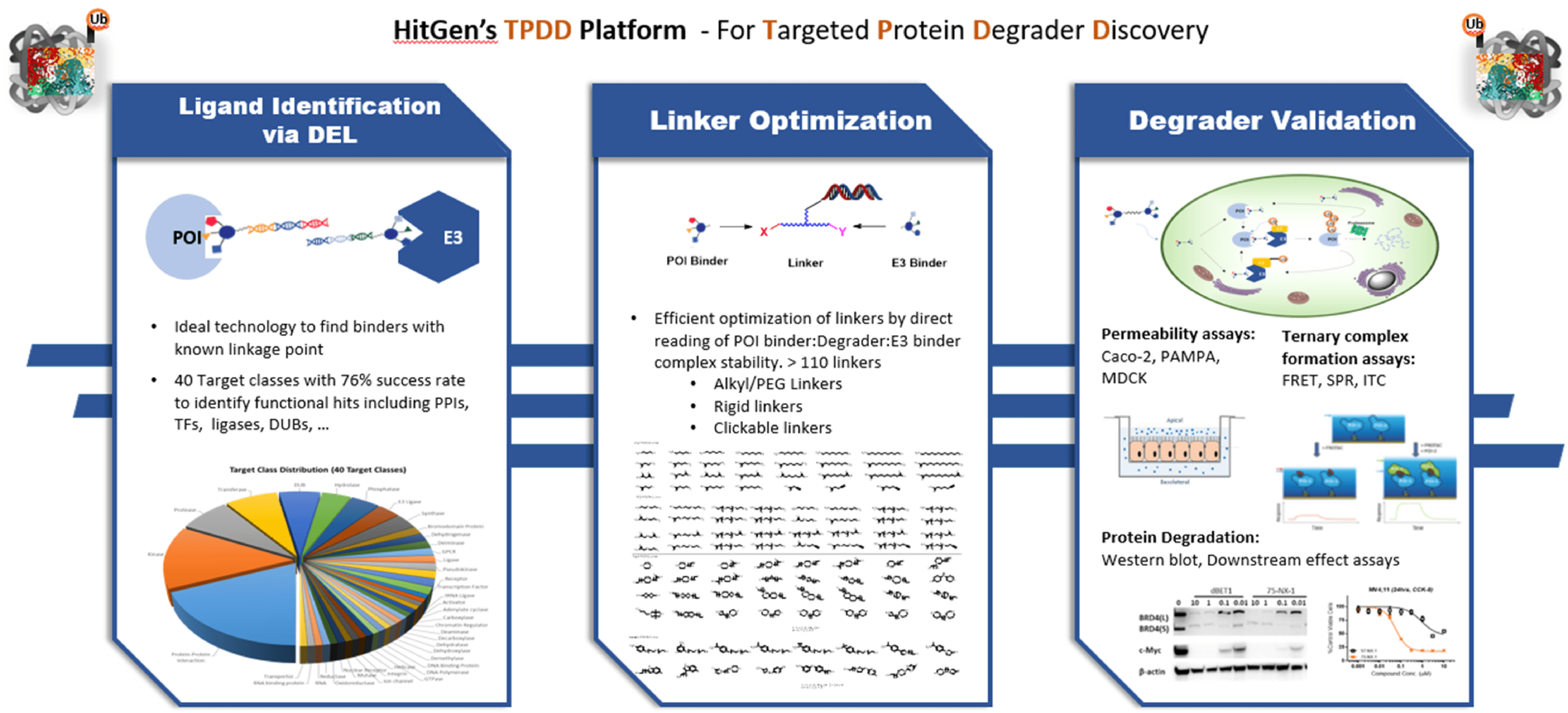
Combined with a series of biochemical and biophysical assays for degradation validation, HitGen opens the TPDP-Targeted Protein Degradation Platform- to embrace any potential collaboration opportunities on the new drug discovery paradigm. TPDP includes:
POI Binder Identification
DNA encoded compounds are covalently linked to DNA tags, having similar molecular architecture as PROTAC molecules.
DEL screening, based on affinity selection, is able to identify binders for functional and nonfunctional proteins. The binding affinity ranged from low nM to low μM.
DEL screening is a very efficient and cost-effective approach for ligand finding.
HitGen has screened 41 target classes including PPIs, ligases, Transcription factors, and many other challenging targets.
E3 Ligases and Their Binders
In the E3 ligase family, there are more than 600 ligases reported. However, only very limited number of E3 ligases, such as CRBN, VHL, MDM2, and cIAP, have been understood and explored for protein degradation.
HitGen is working on identifying novel E3 ligases, and expanding the E3 ligase and binder tool box to be used for both external and internal TPD projects.
Linker Optimization
Protein degrader research requires a lot of efforts in linker optimization in the context of bifunctional molecule development. The optimization process is time consuming due to unclear guidance as to which attaching points, ligands, linker type and length would give optimal degradation efficacy. To make the optimization duration shorter and the process more straightforward, HitGen developed two different approaches for project investigation.
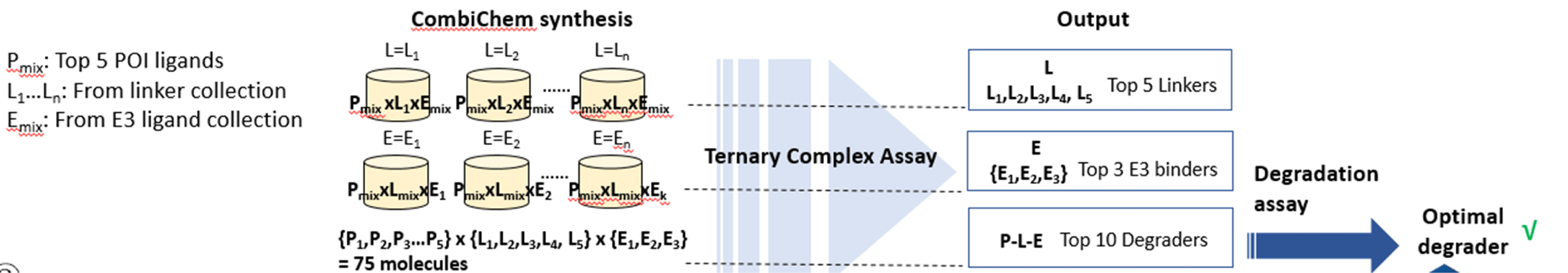
1. CombiChem approach
This approach can be exploited when E3 ligase and binders are not defined. With top POI binders identified from DEL screening (attaching points given), degrader mixture will be synthesized with the combination of multiple of E3 ligase binders, and each mixture will use one specific linker type. Followed by ternary complex assay, the optimal linker can be obtained can synthesized with top POI binders, while varying the E3 ligase binders to identify the top E3 ligands. In the end, degrader molecules will be synthesized to be evaluated with degradation assay such as western blot.
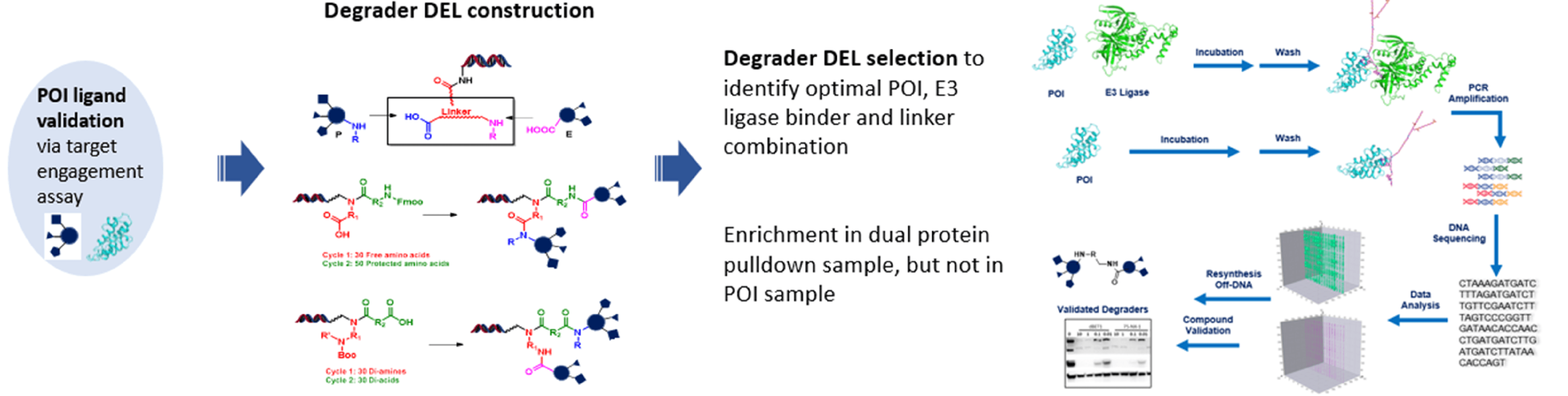
2. DEL Approach
In this approach, both POI binders and E3 ligase binders are given. A DEL library will be constructed to yield different combinations of POI binder-linker-E3 ligase binder, and followed by focused selection the optimal combination will be recommended as a result of ternary complex harvest.
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information