One-stop Technology Platform to Accelerate Drug Discovery and Development
DNA Encoded Library Screening (also known as DEL selection) is an affinity selection process. DEL screening has many advantages over traditional approaches: 1) easily accesses trillions of physically existing compounds; 2) saving in protein/target consumption; 3) short duration in hit identification and validation; 4) more cost effective.
DEL selection principle and strategies for finding functional hits
DEL screening uses very high concentration of targets (μM range) to drive the equilibrium towards the formation of Target-Compound complexes (the direct readout is the abundancy of Target-Compound complex), therefore much lower compound concentration is needed (10^5 per compound or lower) in screening and billions to trillions of compounds can be evaluated in a single vial.
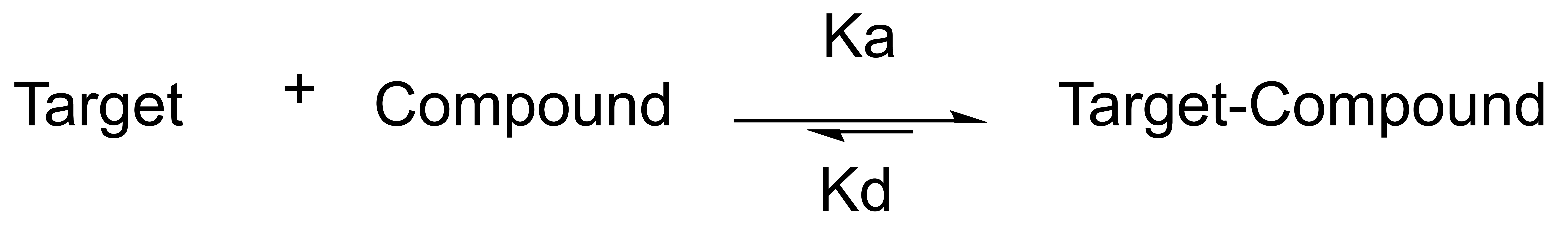
DEL selection uses multiple samples to distinguish compounds with desired function(s) rather than just finding binders. Please read our cases studies for details.
DEL selection target requirements
>90% purity; a clear determination of the multimeric status is recommended
His, biotin, Strep, GST, FLAG and Fc tags are compatible (first 3 tags or dual/multiple tags are preferred)
Tag should not impair target activity and functionality
The required protein amount is dependent on the selection group design. For a typical 50 KD protein with 6 groups in selection plan, 1 mg is sufficient
For cell surface and cellular targets, please refer the following publications for target requirement: ACS Comb Sci. 2015 Dec 14;17(12): 722-31; J Am Chem Soc. 2019 Oct 30;141(43):17057-17061; Nat Chem. 2021 Jan;13(1):77-88.
DEL selection against multiple DELs (DEL pooling)
At HitGen, hundreds of libraries are pooled to one or several DEL packages for target DEL selection, while considering target usage efficiency, target type, DEL type, optimal DEL population as well as DEL solubility. HitGen offers highly efficient and flexible DEL pooling strategies with assistance of automatic or semi-automatic instrumentation. In addition, highly customized pooling strategies are also available.
DEL selection and hit identification process
Target activity confirmation and pre-selection studies
Pre-Selection studies include a careful assessment regarding the target activity confirmation and selection feasibility to ensure DEL selection success and minimize the chance of false positive hits. A thorough assessment of each component will be conducted, aiming to accomplish a well-characterized selection buffer in which the target would behave in the same way or at least similarly to its original buffer. Pre-selection studies will help build confidence for the initiation of DEL selection by acknowledging that the target shows good activity under selection conditions, and the immobilization will not impede the binding between the target and DELs.
DEL selection processes for different DEL types
Regardless of the library type, DEL selection involves the following general steps: target-library incubation, unbound molecule removal, and bound molecule recovery. Each step is of great importance in achieving potential hits. HitGen provides various DEL selection options involving different types of libraries, and each type of selection follows distinct procedures. Next-Generation Sequencing (NGS) is utilized for the sequencing process. In order to achieve high data usage efficiency, a certain range of copy number in each sample is necessary. We usually need to decide where we should stop the multi-round selection by monitoring copy number of the elution with Quantitative Real-time PCR (qPCR). A classical DEL selection scheme is shown below, please read more for Covalent DEL selection, protein degrader DEL selection, and Fragment DEL selection.
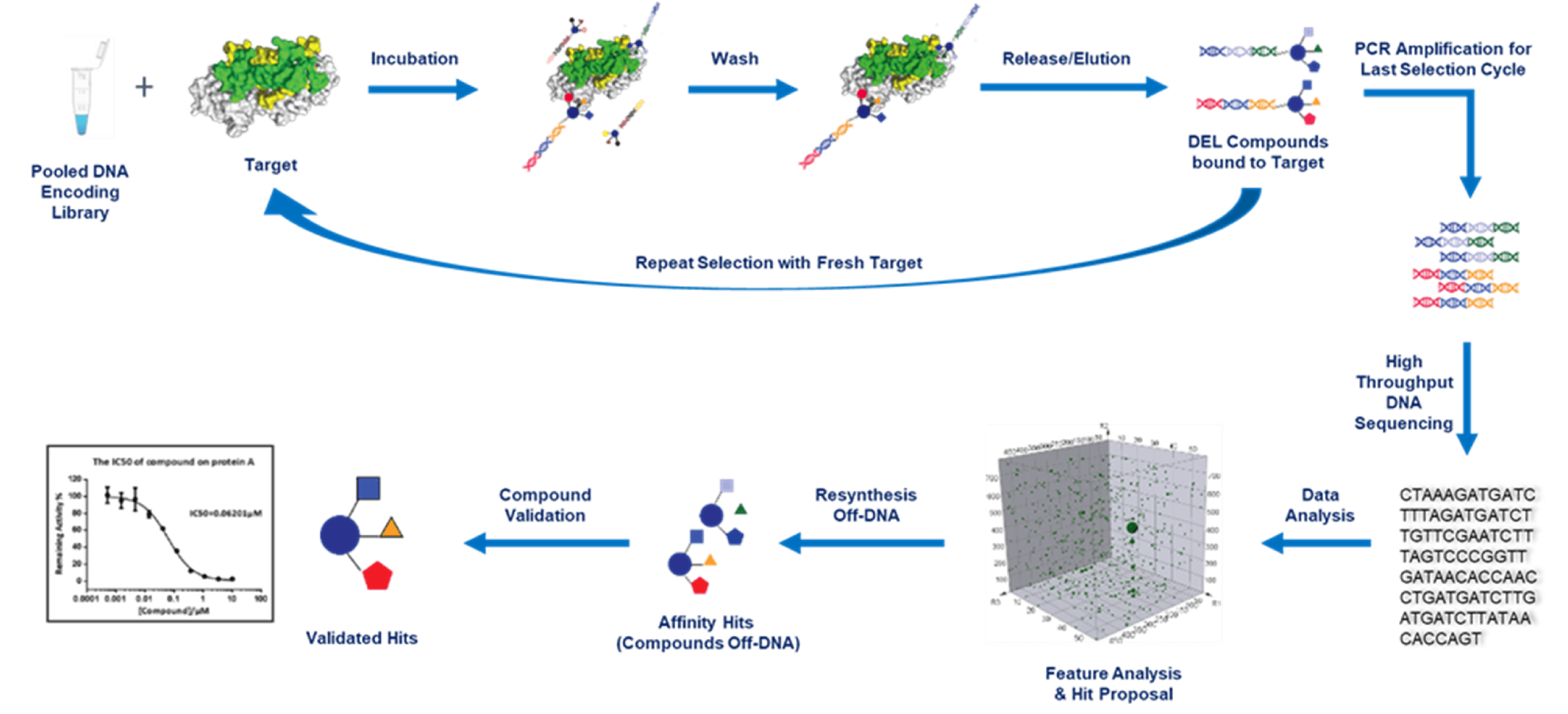
In most cases, DEL selection at HitGen is performed with automated sample purification system (KingFisher Due prime and KingFisher Flex). For detailed selection automation, please refer to a well written paper of DEL selection using Kingfisher: Nat Protoc. 2016 Apr;11(4):764-80.
DEL selection compound input
DEL input refers to the copy number of individual DEL molecule in the selection. We have a systematic study on the relationship between DEL input and selection performance, please refer to our publication for more details (SLAS Discov. 2020 Jun;25(5):523-529).
PCR and DNA sequencing
At HitGen, two Illumina sequencing instruments, Hiseq2500 and NovaSeq6000 are used in DNA sequencing. Based on needs, 6 different types of flowcells are used for the sequencing of DEL selection PCR amplified products. This offers flexibility in sequencing capacities from 115M to 6400M reads and robust sequencing quality with average Q30≥90%. In house DNA sequencing provides zero delay in the analysis of DEL selection results and typically the sequencing is comleted in one day. We have a patented PCR method (patent# 201811151077.3) to prepare the sequencing libraries of DEL selection samples.
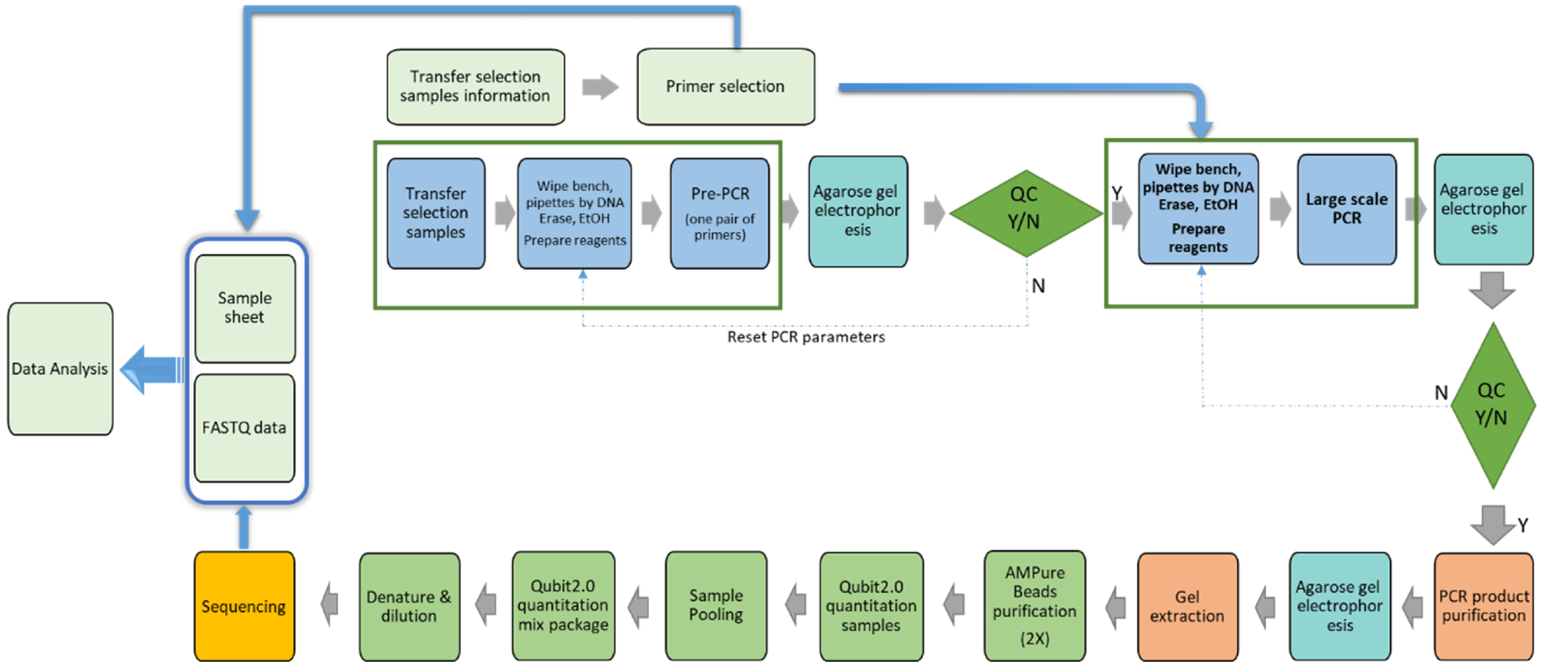
Sequencing data deconvolution/affinity hits identification algorithm
Sequencing data deconvolution is the process of converting the selection readout (in the form of DNA sequences) to chemical compounds that these DNA tags encode including the specific scaffold, building blocks and library chemistry employed. PolyO (HitGen’s proprietary hits identification algorithm) is highly a sensitive method to identify feature *(* feature refers to a group of compounds sharing common scaffold and/or building block(s); typically it is represented as a line in the cubic view) enrichment from high-throughput sequencing data. These features are then analyzed by comparison of samples and classified to different scenarios for further analysis using Data Warrior.
For each DEL selection, we normally examine hundreds of DELs with enriched signals. In order to improve the speed of the data processing and make timely data-driven decisions, we have built the DELT data analysis automation platform for data auto processing and reporting. This automation system is able to present the selection data in roughly one day right after the completion of sequencing.
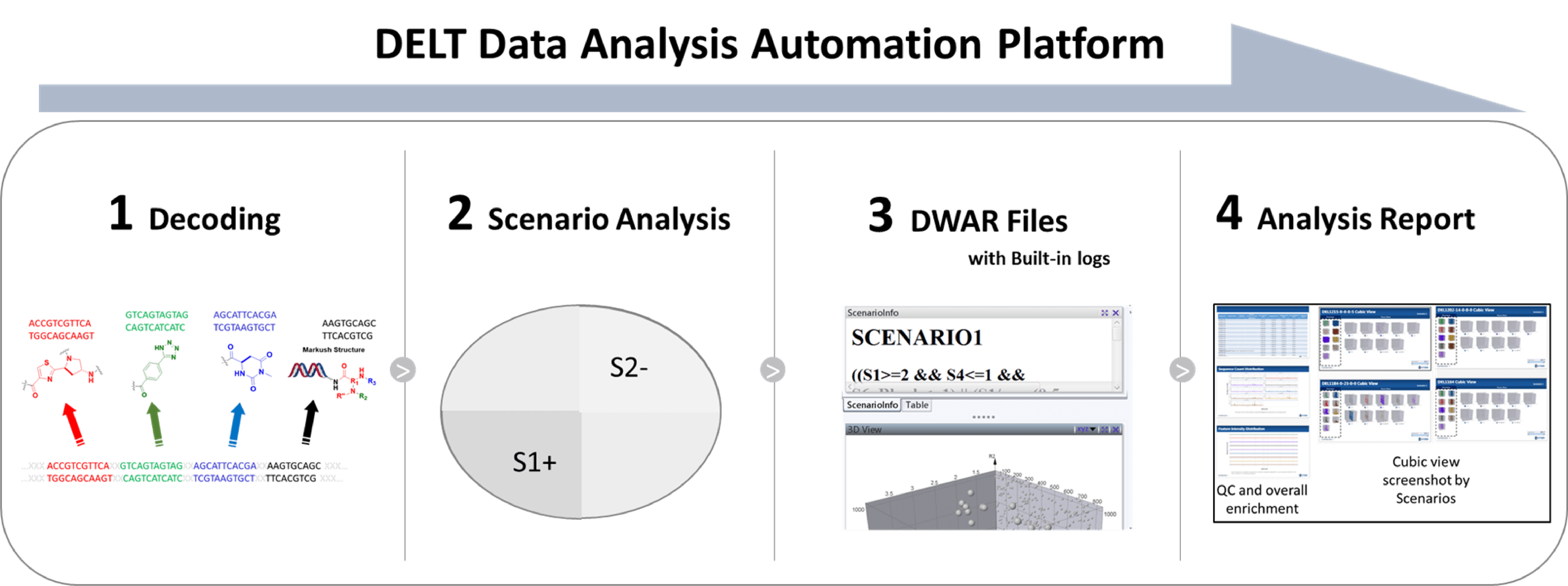
Hits nomination and off-DNA synthesis
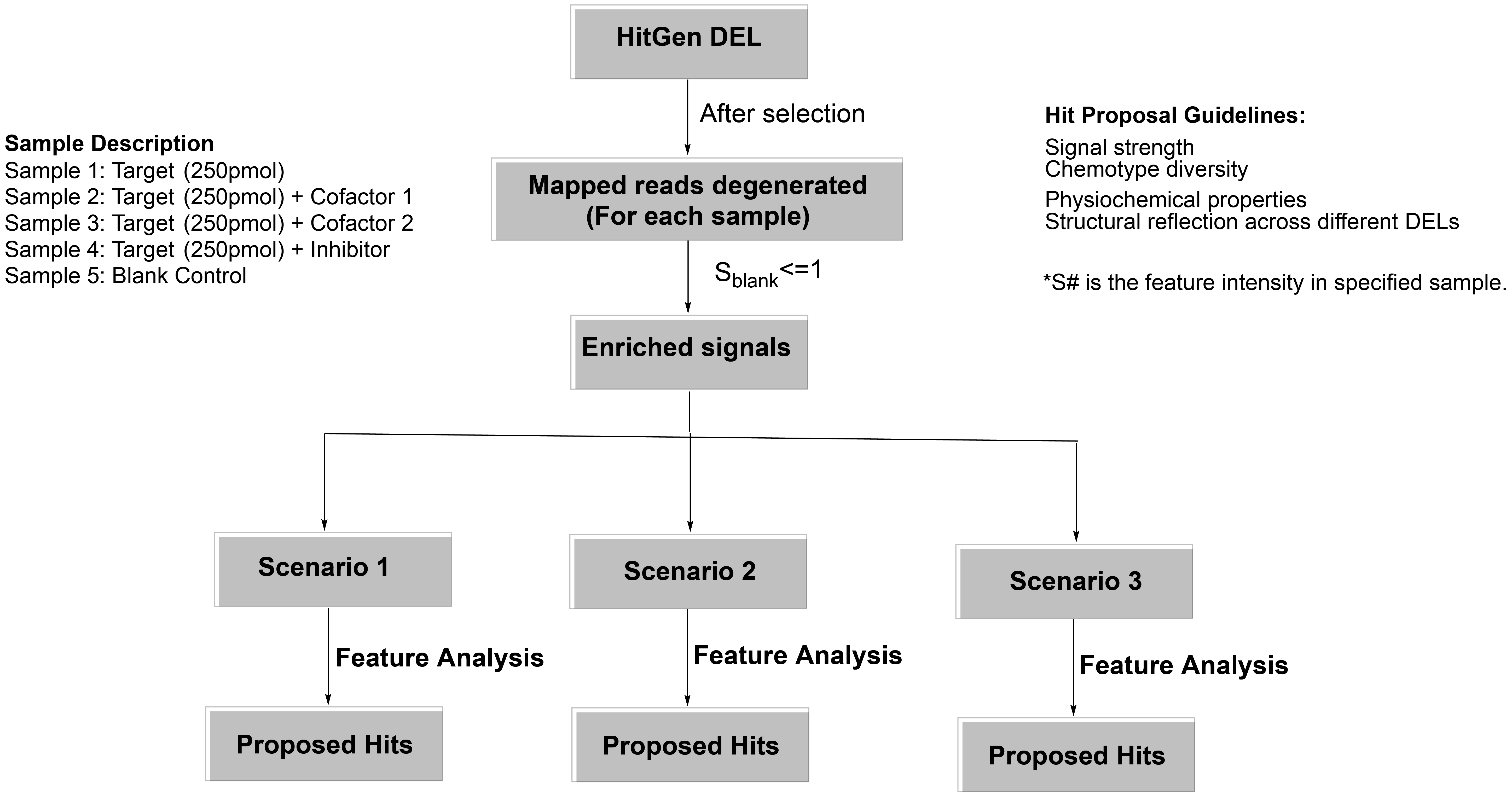
For hit proposal, we generally have the following considerations: 1) signal strength (sequence count, feature intensity), 2) chemotype diversity, 3) physchem properties, 4) structural relationship across different DELs and mechanism of action (signal comparison between samples). A representative hit proposal workflow is shown below.
HitGen is capable of industry leading synthesis with experienced chemists that have worked on over hundreds of projects and advanced equipment, instruments. Typically, 5 mg is offered for every compound and for up to 100 compounds, with 6~8 weeks lead time. Gram-scale of synthesis is also available as needed.
Biophysical and biochemical assays for hits validation
ASMS for feature compound identification
Affinity selection-mass spectrometry (ASMS) is used in on-DNA validation in order to select possible binders with high confirmation rate. This approach takes both products and byproducts into consideration by mimicking the reactions during DEL production. We are also exploring different kinds of cleavable linkers, which will maintain the same conditions in DEL production but also avoid affections from their DNA tags during affinity selection.
DEL results driven hit-2-lead optimization
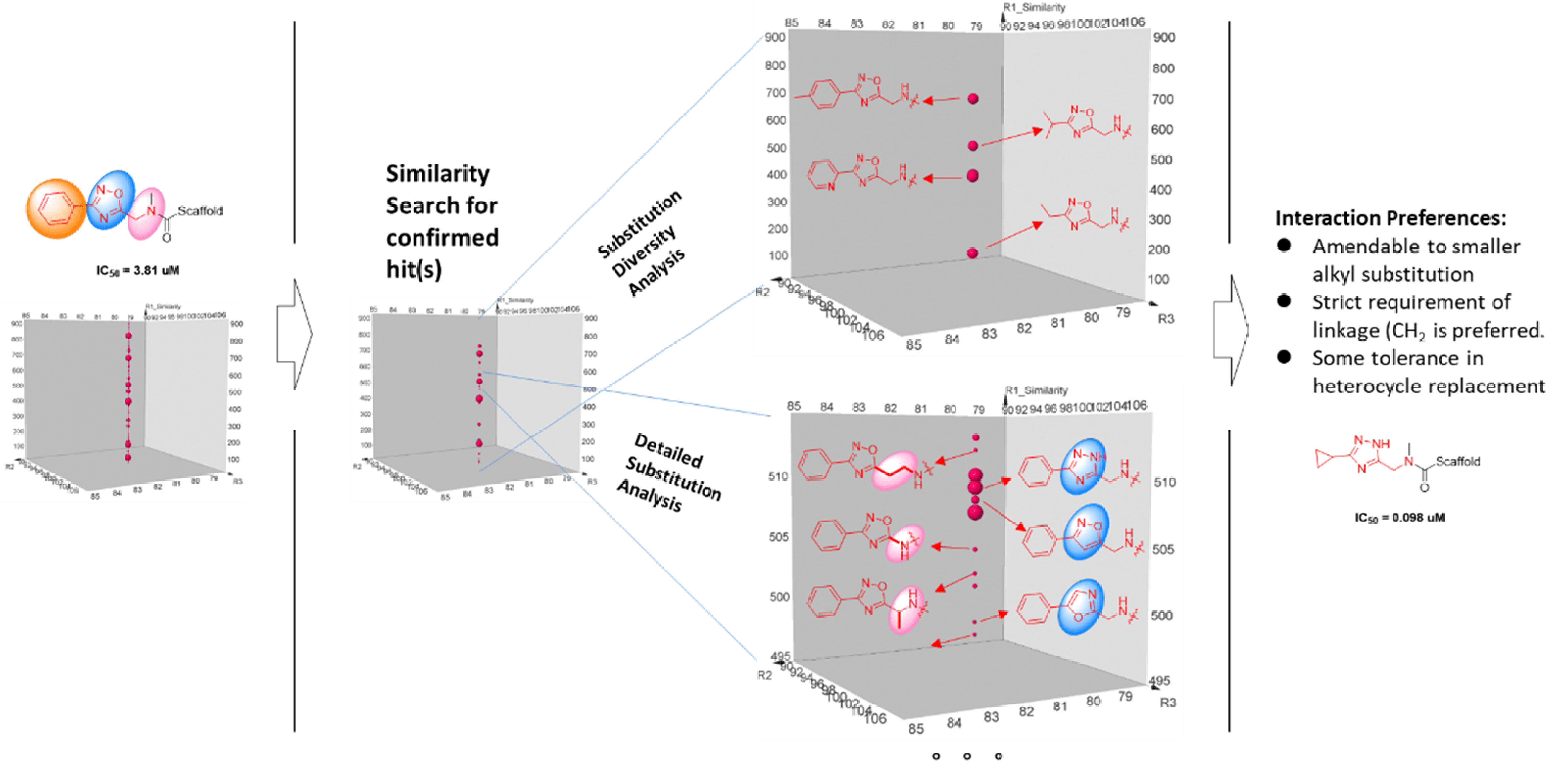
The tremendous DEL screening information is a great resource for further probing target’s pocket and provide compound optimization directions. Structure-Signal Relationship analysis is largely conducted on the same feature which entails the compounds that involved in the highly similar, if not the same, chemistry, the difference of sequence count should be mostly due to the result of natural selection of the target. With the per stated premises, sequence count positivity correlates to bioactivity. A representative Structure-signal Relationship Analysis is shown below.
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information