One-stop Technology Platform to Accelerate Drug Discovery and Development
Identification of Transferase NAA50 Inhibitors by DEL Selection
(Collaboration Project with Pfizer, ACS Med. Chem. Lett. 2020, 11, 1175−1184)
Protein NAA50 and its function
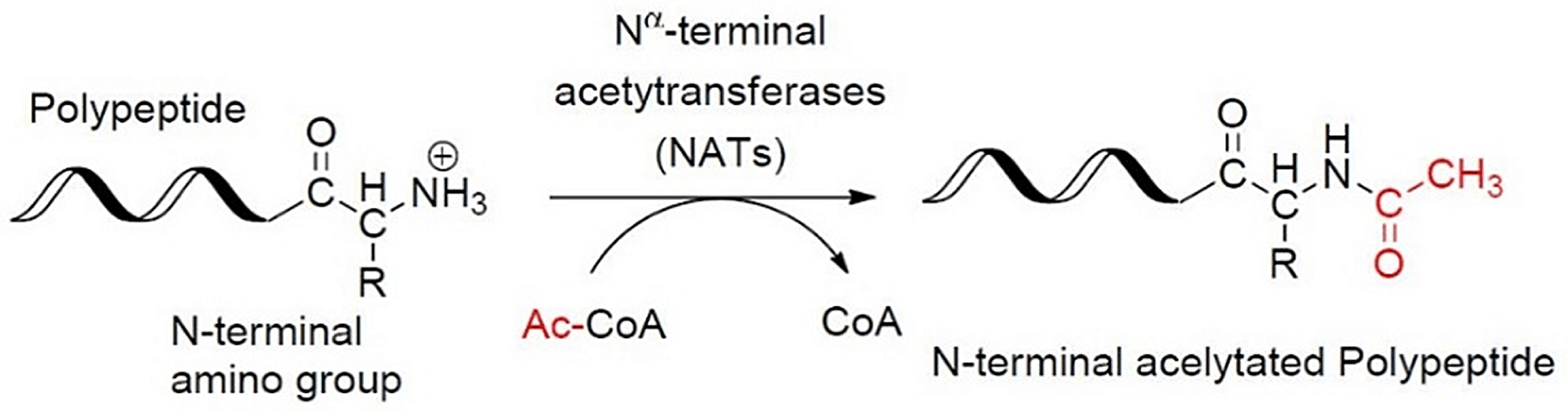
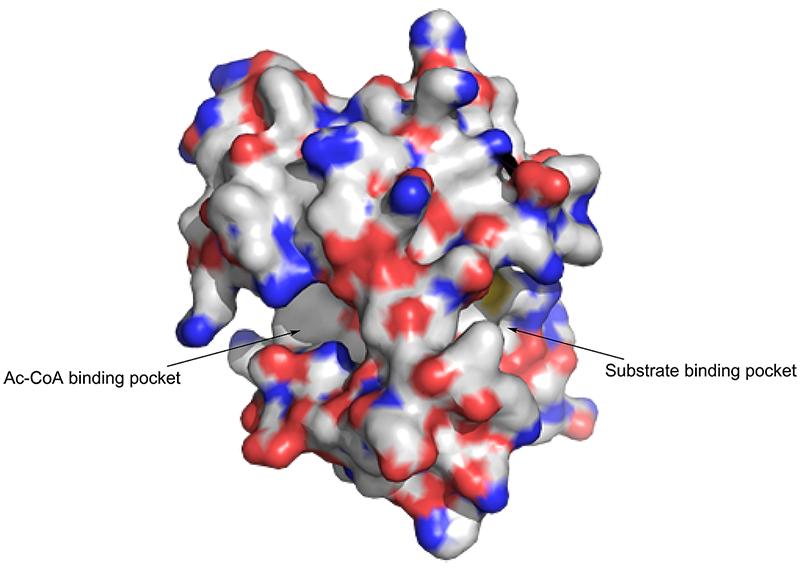
The N-terminal-acetylation of a protein can affect its nuclear import and export and can also act as a degradation signal to control the protein’s cellular stability. The Nα-terminal acetyltransferase (Naa50) enzyme is a member of the Nα-terminal acetyltransferase NAT protein family. It coexists with Naa10 and Naa15 in the NatE complex and is responsible for the enzymatic function of the complex. Naa50 is also found to be essential for normal sister chromatid cohesion and chromosome condensation. Therefore, an inhibitor of the Naa50 enzyme might have therapeutic applications in oncology indications. The enzymatic catalysis and protein structure are shown below.
NAA50 known inhibitor and objective of DEL selection
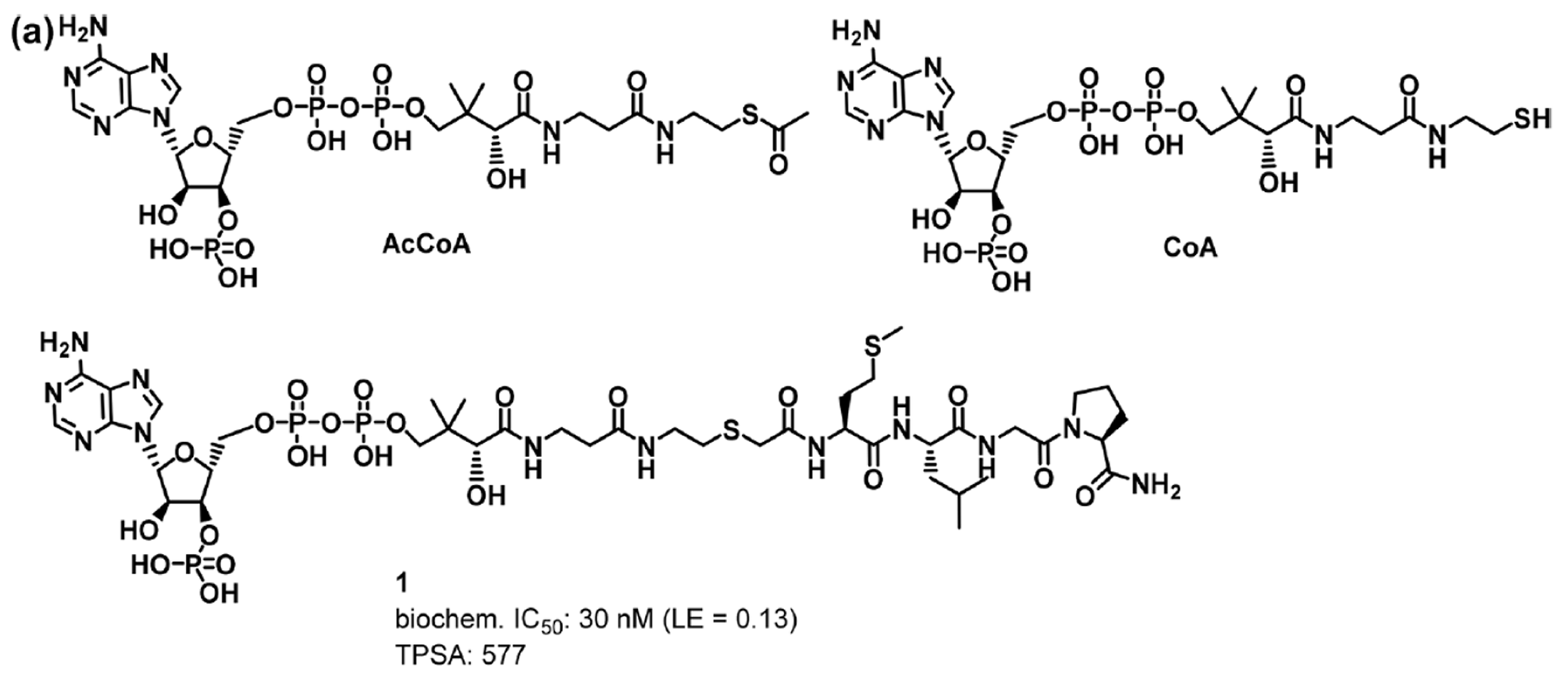
Compound 1 is designed by studies of the NAA50 biochemical mechanism which indicated formation of a ternary complex between the AcCoA cofactor, an appropriate protein substrate (tetra-peptide MLGP), and the enzyme. Although compound 1 is a potent Naa50 inhibitor, the molecule is not particularly efficient due to its large molecular weight (ligand efficiency (LE)10 = 0.13). In addition, its high molecular weight (MW = 1223) and high polarity (tPSA = 577 and cLogP = −4.1) likely prevents facile permeability across cell membranes and may thus compromise the use of the molecule as a robust in vitro tool compound.
The DEL selection is to identify potent and selective NAA50 inhibitors with improved physiochemical properties relative to compound 1 (i.e., reduced molecular weight and tPSA; increased logD).
DEL selection plan
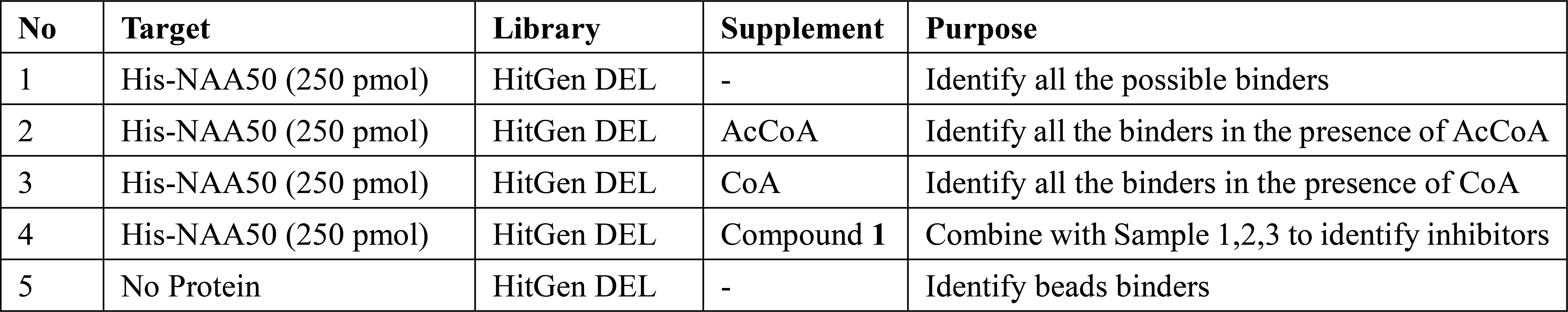
In a typical DEL selection plan, we set up 3 samples (1. protein alone, Apo NAA50 in this case; 2. protein and inhibitor in saturated concentration; 3. Blank control). However, during the studies of NAA50, we realized that NAA50 protein involves conformational change in the catalytical process. By comparison of selection results of Sample 1 and 2 aforementioned, we are not able to effectively identify inhibitors. Consequently, we included two additional samples by addition of AcCoA and CoA, seeking for compounds binding to the transition state. The final Selection Plan are summarized below.
Representative DEL selection results
DEL selection is typically presented in a cubic layout, where the axis represents the corresponding building block and bubble size represents the sequence count of each compound. If the compounds have more engagement to the protein (high binding affinity or slow off-rate), the sequence counts will be presented as bigger bubbles. As we can see from the first four samples (blank sample 5 is now shown), Apo NAA50, Apo NAA50 + AcCoA, Apo NAA50 + CoA and Apo NAA50 + Compound 1 (also called Bi-subi) resulted different enrichment pattern, proving that NAA50 has conformational changes during the catalysis. Selection of compounds binding to all the protein stages and competing with Compound 1 obviously will yield NAA50 inhibitors. Such a selected compound structure is shown in the figure below, where the compound is built in the DEL as a racemic mixture.
The selected compound DEL951-34-888-1668 is resynthesized for validation.
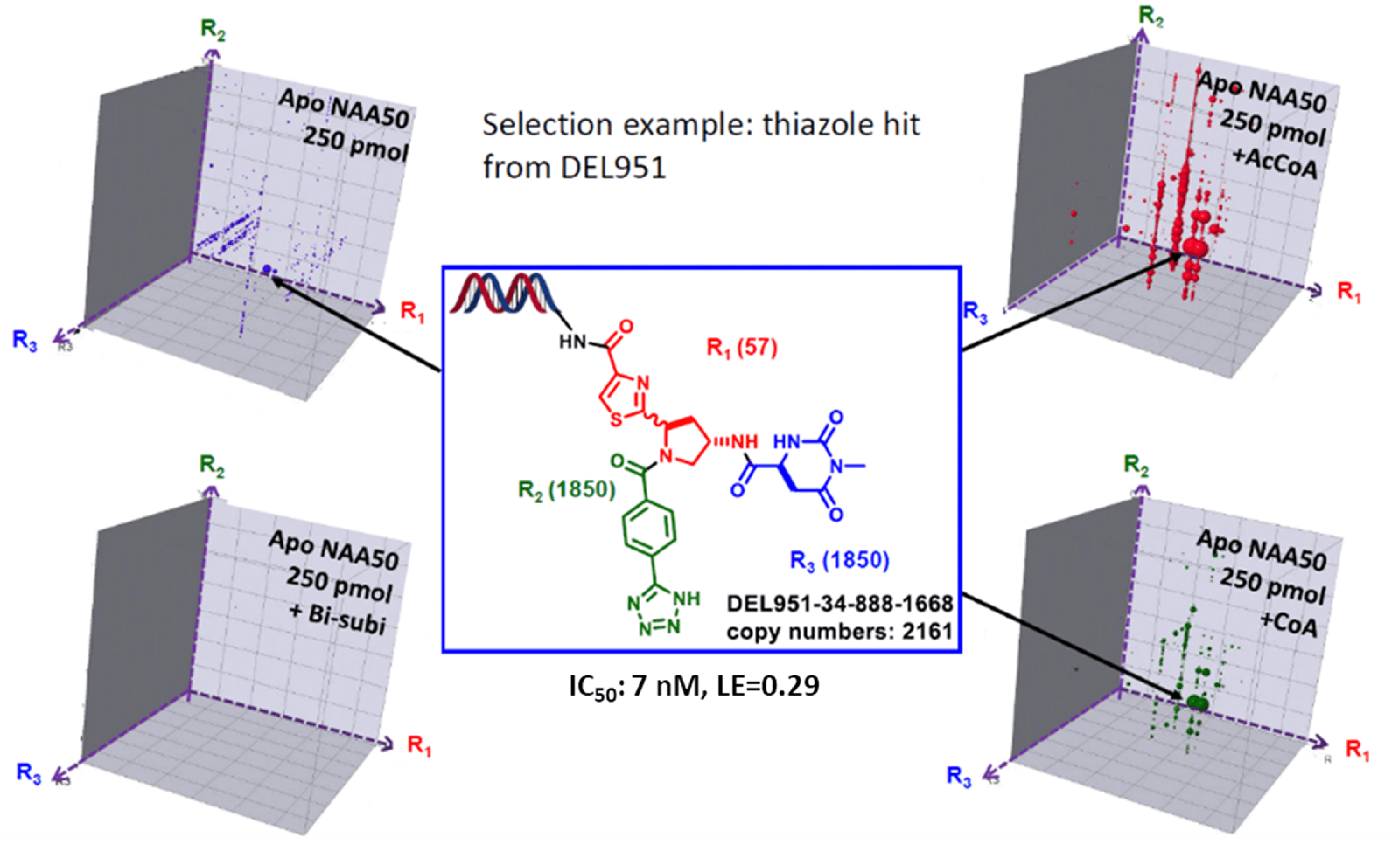
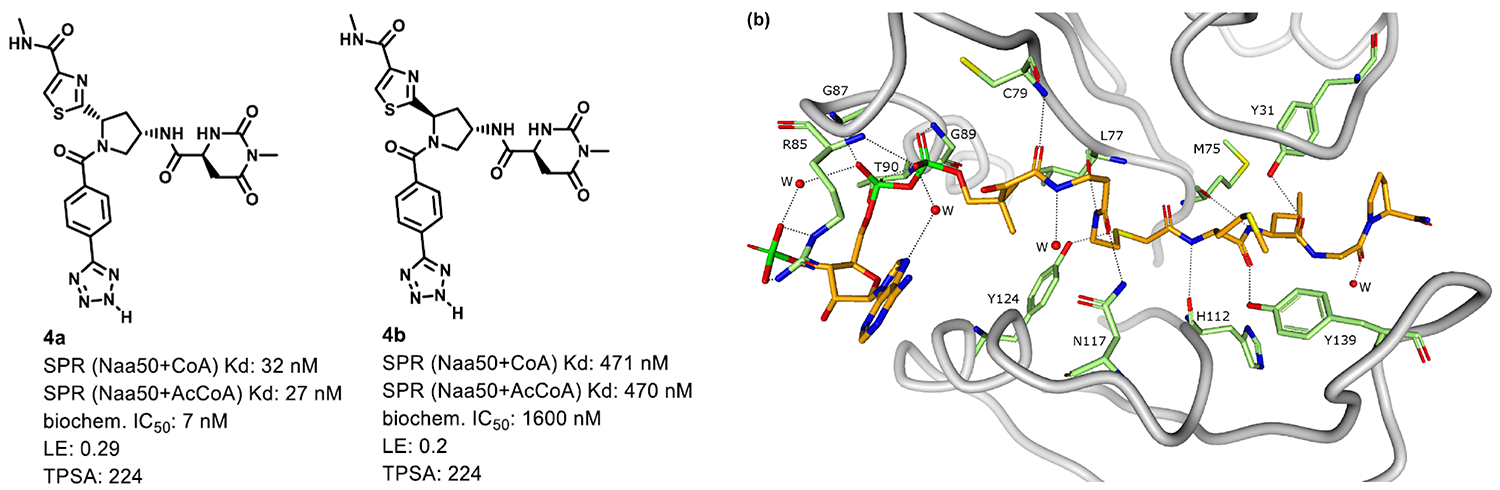
DEL compound validation
The two isomers of the selected compound DEL951-34-888-1668 were synthesized and tested by SPR in the presence of CoA and AcCoA and biochemical assay. The chiral isomer 4a has been found as a very potent inhibitor with improved MW, Ligand Efficiency, and tPSA. The interaction of compound 4a and NAA50 has been further confirmed by co-crystal structure by Pfizer (pdb code: 6WFN).
Novel BRD4 degrader discovery using DEL technology
Advantages of using DEL selection for protein degrader discovery
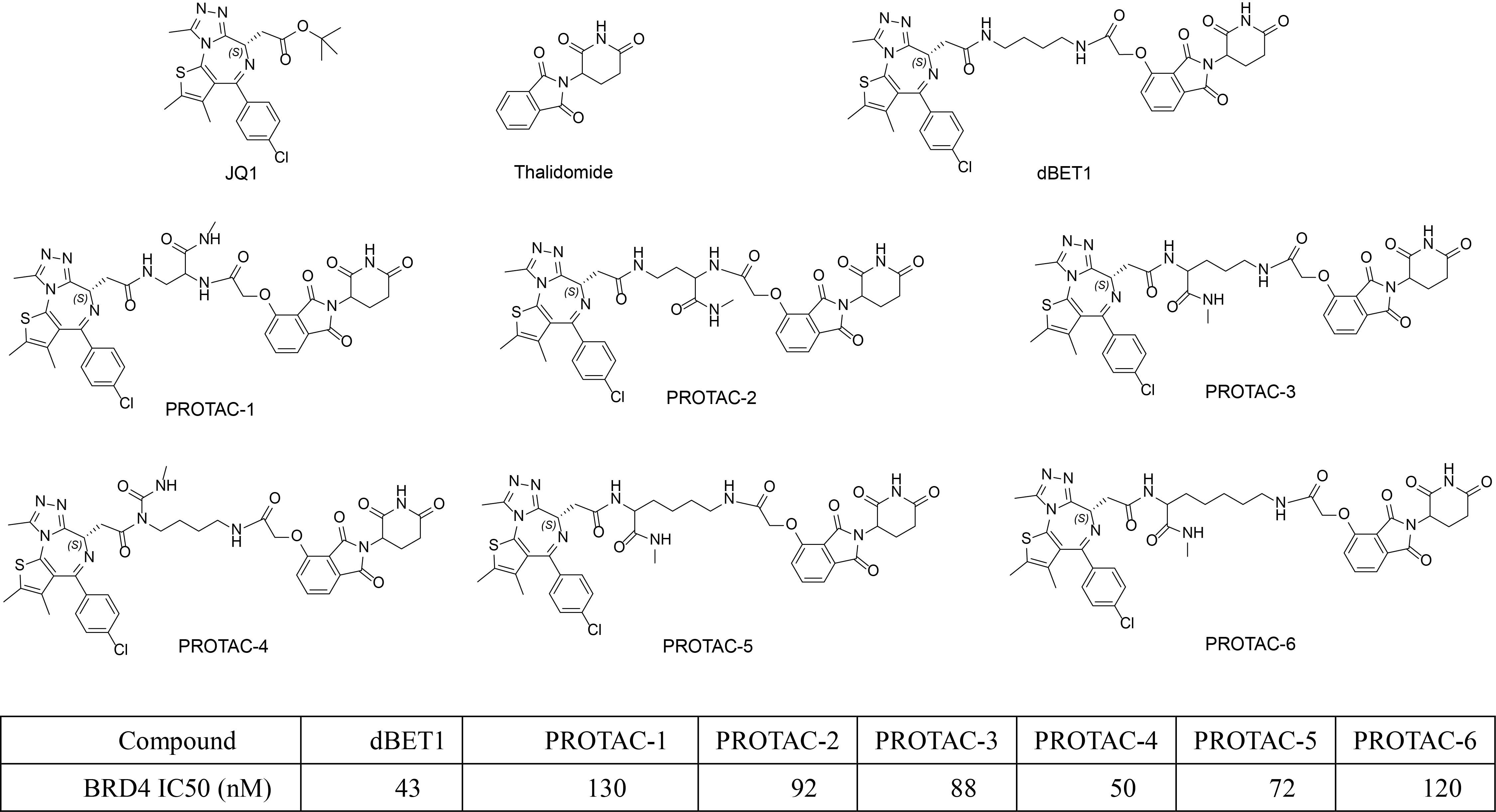
DEL selection is an affinity-based approach to recognize compounds interacting with the target, including the compounds modulating target function(s) or simply binding to the protein. The architecture of a DNA Encoded Compound is very similar to proteolysis targeting chimeras (PROTAC) as shown in the following figure. Both DEL compound and PROTAC molecule require covalent linkage of two molecules with known attachment points that have minimal impact to the binding.
More importantly, DEL selection is able to identify affinity binders for Protein Of Interest (POI) and E3 ligases, therefore it has advantages in creating stronger intellectual properties and exploring therapeutic benefit from novel E3 ligase(s) by their tissue distribution differences.
In a traditional DEL selection, where target is seeking for compound, the direct readout of DEL Selection is the absolute number of DNA sequences (ideally prepotentional to number of compounds). In principle, when both POI and E3 ligase are used in the DEL selection, we are able to identify compounds binding to both proteins, using POI and/or E3 ligase as control(s).
POI-compound-E3 ligase ternary complex stability is the key for protein degradation
In order to prove the above theory, we have used JQ1 as BRD4 binder, thalidomide as CRBN binder, linkers with different lengths to test whether we are able to distinguish the ternary binding stability. Specifically, six PROTAC molecules and dBET1 (a well-known BRD4 degrader, Biochem Biophys Res Commun. 2018, 497(1):410-415.) were made and their IC50 values were measured. From the IC50, we are not able to know which one(s) bind to both BRD4 and CRBN to form more stable complex.
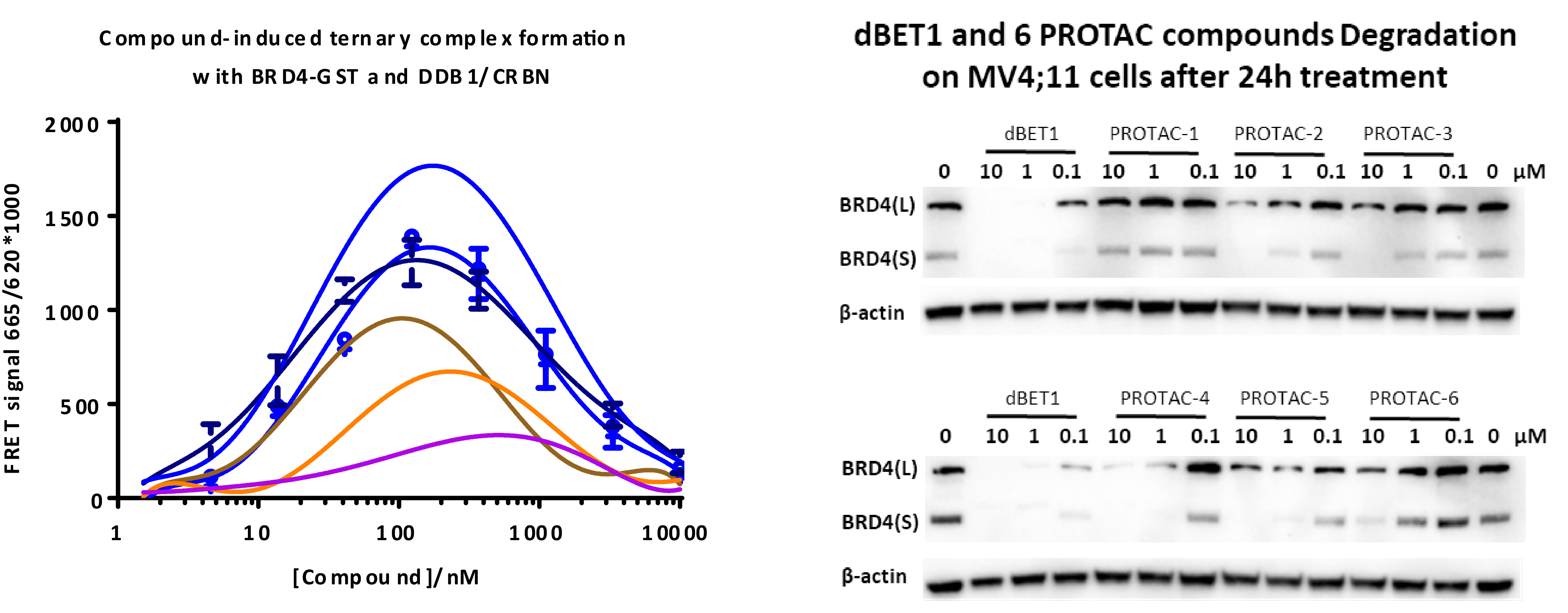
Then these 6 PROTAC molecules and dBET1 were evaluated with a FRET based ternary complex formation assay, where they clearly showed different capabilities in ternary complex formation. BRD4 degradation experiment in MV4;11 confirmed the differential degradation of these 6 PROTAC molecules.
DEL selection approach positively correlates to complex stability
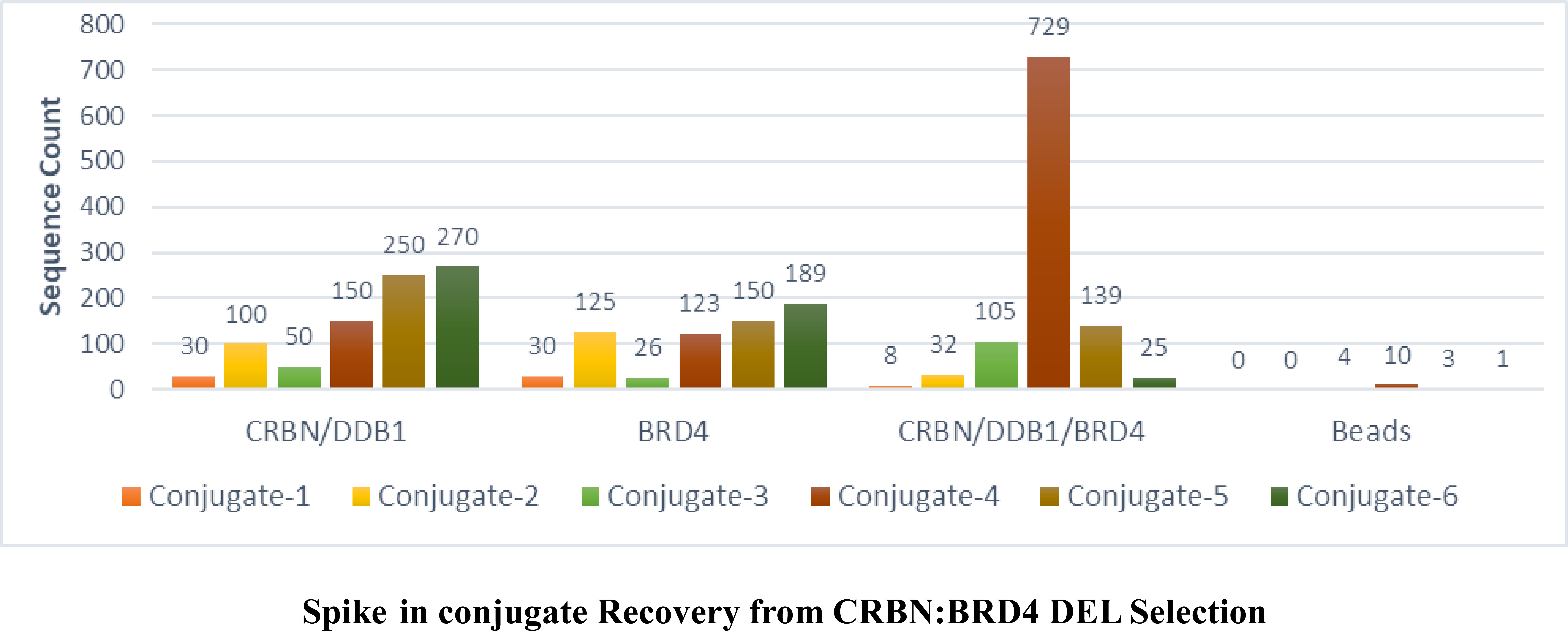
In order to confirm that DEL selection is able to distinguish complex stability, we made 6 corresponding PROTAC-DNA conjugates to see if different recovery can be observed from a DEL selection. In the “Conjugate Recovery Experiment”, Conjugate 1~6 were pooled into DEL library; a DEL selection process was conducted and the corresponding DNA sequence counts were compared (DEL selection please refer to “PROTAC DEL Selection”). Conjugate-4 is readily recognized as the optimal degrader molecule since it has significantly higher sequencing count, which is corresponding to number of compounds binding to CRBN-BRD4 complex. The length is consistent to optimal linker length in dBET1. On the other hand, recovered DNA sequences from CRBN and BRD4 samples did not give the correct trends.
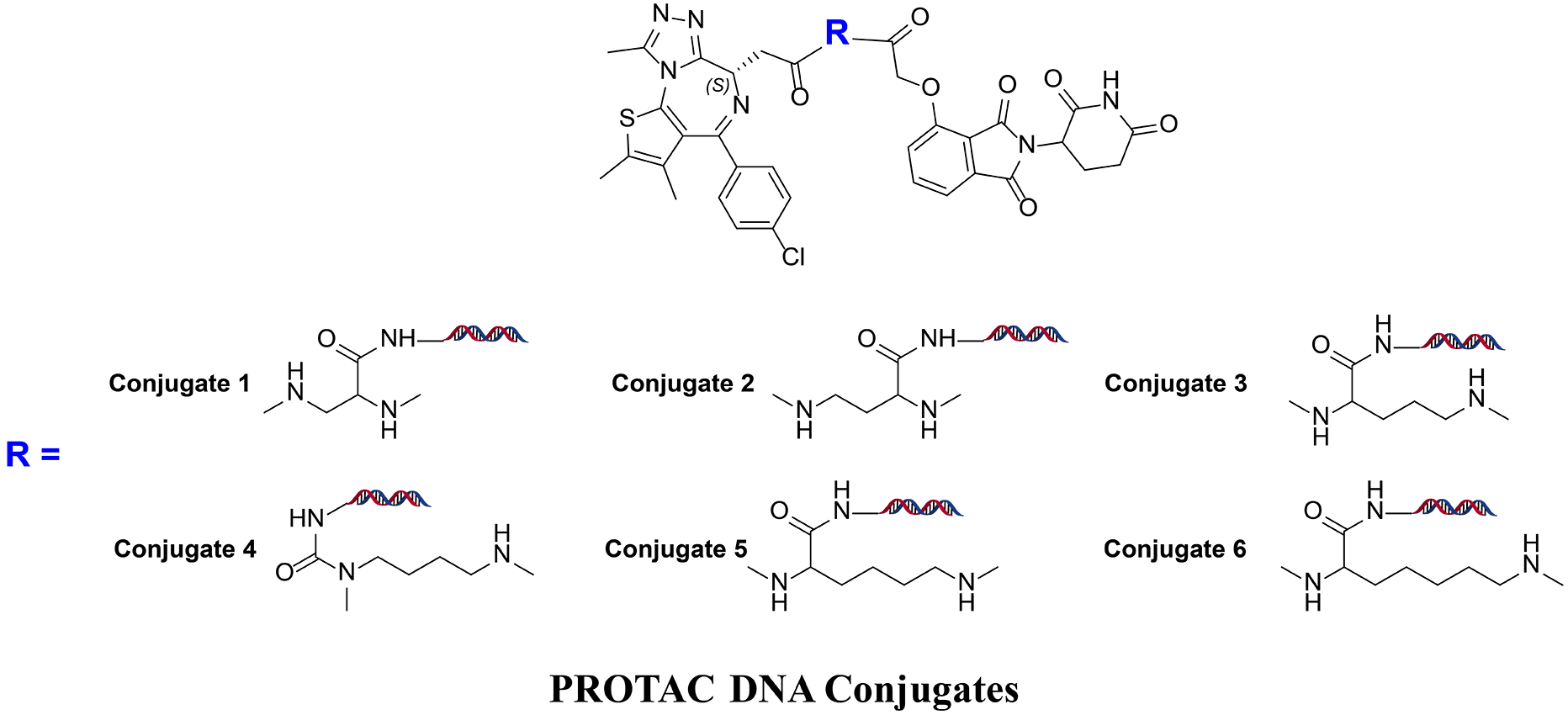
Discovery of novel BRD4 degraders using HitGen PROTAC DELs
HitGen PROTAC DELs include E3 ligase binders and diverse chemical structures. Below is one of the PROTAC DEL general structures, where thalidomide is to target CRBN and R1, R2, R3 are linker and POI binding variation, respectively.
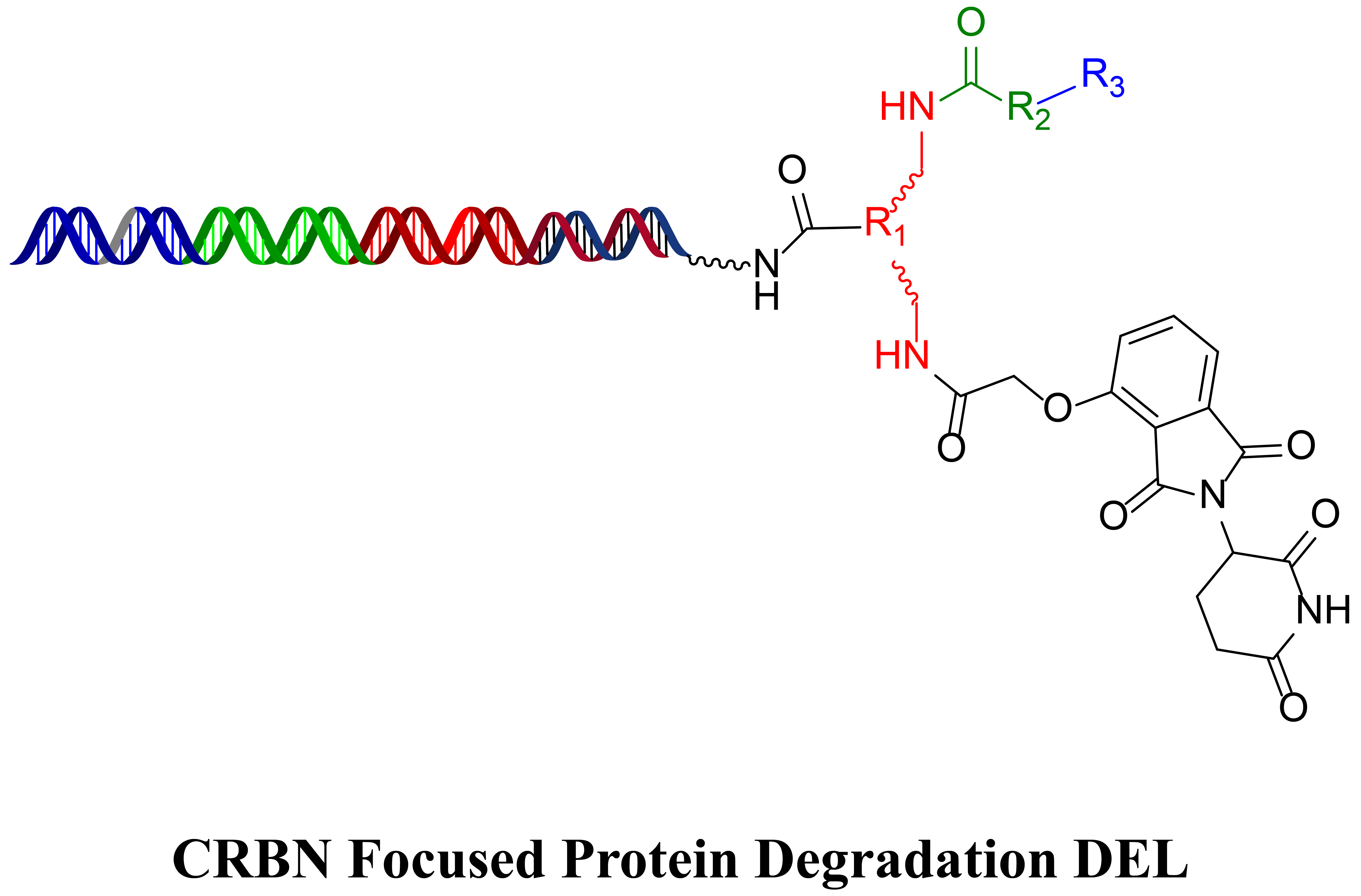
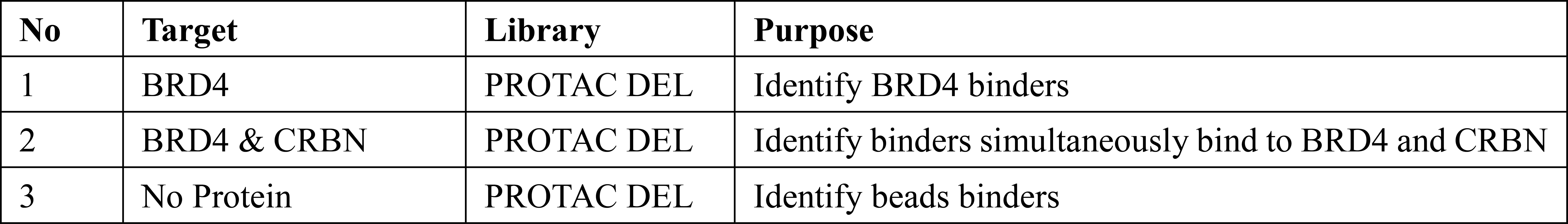
PROTAC DEL selection plan and objectives
For this specific DEL, thalidomide has been approved clearly to bind CRBN, therefore DEL Selection against CRBN alone was not included. The Selection Plan is outlined below.
The aim is to identify any novel compounds binding to both BRD4 and CRBN over BRD4, subsequently proving that the PROTAC molecules with strong signals in Sample 2 result better BRD4 degradation.
PROTAC DEL selection results
For clarity, a specific plane with given R3 in the cubic presentation of DEL compound enrichment was compared between sample 1 and sample 2 (Blank sample not shown).
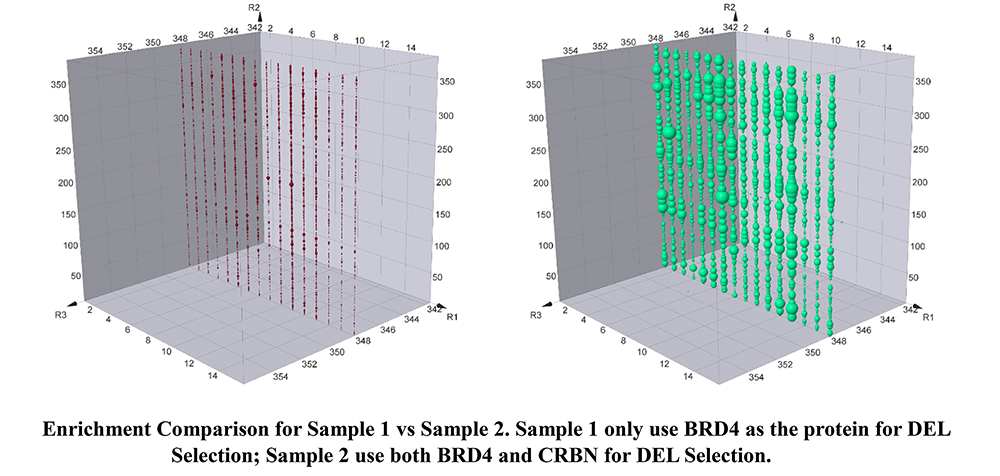
Plane R3=348 clearly suggests that many compounds on this plane are preferentially bind to both proteins (Sample 2) rather than just BRD4 (Sample 1). Meanwhile, there are also many compounds bind to both samples similarly. All the compound structures for BRD4 binding are different from JQ1 or any reported BRD4 binders. In order to compare the structure similarity, tarnimoto similarity is used to describe how different they are. When Tarnimoto similarity is greater than 0.8, two molecules are considered as similar. The representative compounds are selected and their tarnimoto similarity scores are indicated as below.
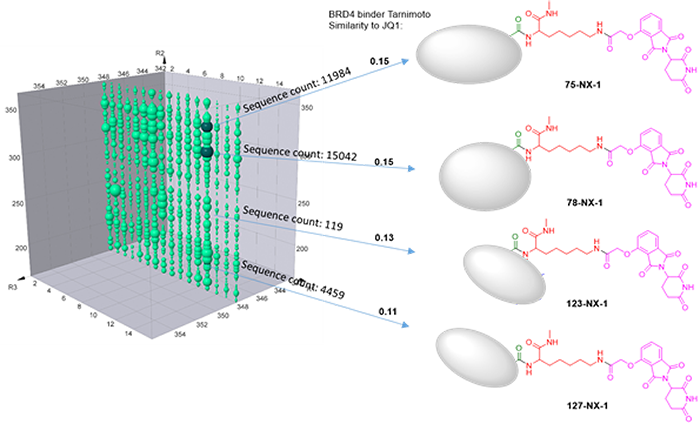
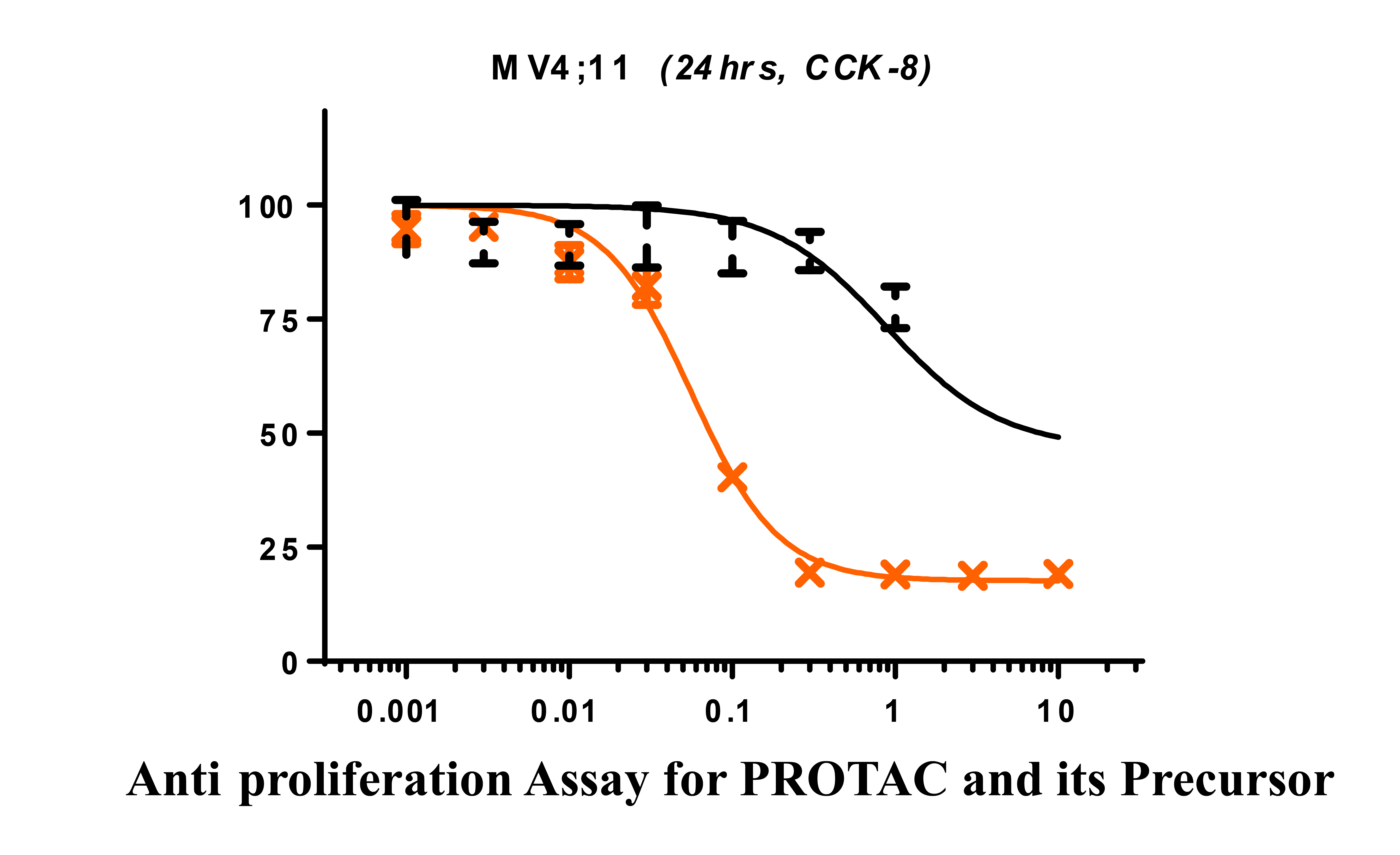
Presumably, the compounds with bigger bubbles (higher sequencing counts) form more stable complexes than that with smaller bubbles. In order to prove this, we have selected 4 representative compounds with different sequence counts in sample 2 (11984, 15042, 119, 4459) for Off-DNA synthesis and protein degradation evaluation (Western Blot). These 4 compounds are named as 75-NX-1, 78-NX-1, 123-NX-1, and 127-NX-1, respectively. Using dBET1 as a reference, compound 75-NX-1, 78-NX-1 with higher sequencing counts observed very similar BRD4 degradation in MV4;11 cell; whereas compound 123-NX-1 almost did not observe BRD4 degradation. Compound 127-NX-1 did not show BRD4 degradation either.
PROTAC compound 75-NX-1 and its BRD4 binding part (57-NX-1, structure not shown) are evaluated in anti-proliferation assay in MV4;11 (CCK-8), the results showed that compound 75-NX-1 has much higher activity than 57-NX-1 (Compound 57-NX-1 was found as a BRD4 inhibitor).
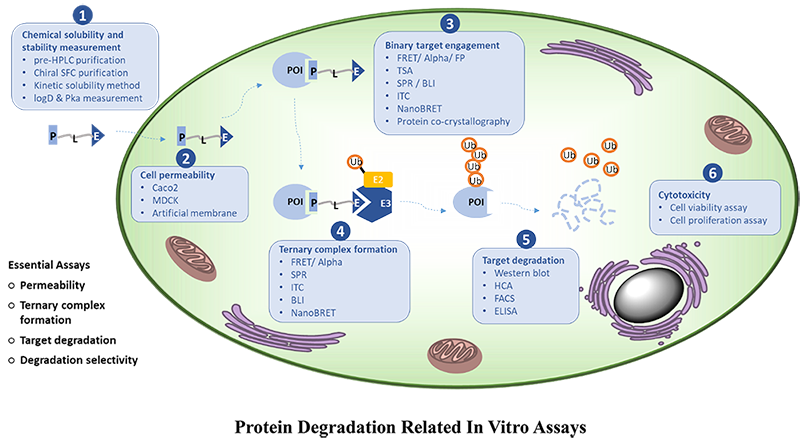
Taken the above together, it suggests that DEL selection is an effective approach to discover or optimize PROTAC compounds to find the most stable POI:Compound:E3 ligase complexes which consequently will have higher chance to achieve protein degradation in cell. We are fully aware that there are additional factors could affect the protein degradation such as PROTAC compound solubility, lipophilicity, orientation and etc (Figure 1), but there is no doubt that DEL selection approach provides a very effective tool for ternary complex formation (an essential step for protein degrader optimization).
We use cookies to provide a better web experience.
By using our site, you acknowledge our use of cookies and please read our Cookie Notice for
More information